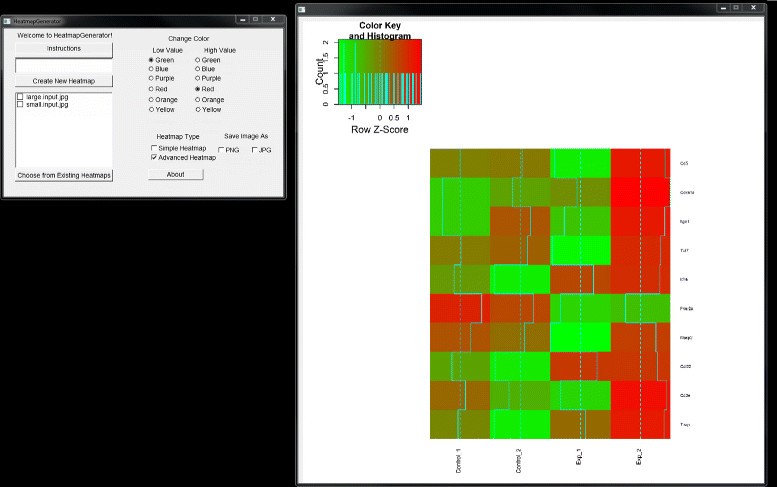
Heatmapgenerator Tutorial For Mac
This is a data styling option in eSpatial. Plot data, then create an intensity/heat map based on pin density or on particular values associated with pins (e.g., sales volumes per year, etc.). Heat Map Tracker records visitor clicks, helps you analyze visitor behavior on your web pages, improves conversion, and helps improve your website results. The ability to understand visitor behavior is very important to the success of your business online. Learn VLOOKUP, IF and many more excel formulas by doing. Unique Excel Test worksheet for training Excel skills. Stop watching videos. Learn VLOOKUP, IF and many more excel formulas by doing. Skip to Main Content. Windows / Mac Requirements: Min. Excel 2010 Bonus. World Countries Geographic Heat Map Generator.
Background The graphical visualization of gene expression data using heatmaps has become an integral component of modern-day medical research. Heatmaps are used extensively to plot quantitative differences in gene expression levels, such as those measured with RNAseq and microarray experiments, to provide qualitative large-scale views of the transcriptonomic landscape. Creating high-quality heatmaps is a computationally intensive task, often requiring considerable programming experience, particularly for customizing features to a specific dataset at hand. Results We create a graphical user interface (GUI) software package called HeatmapGenerator for Windows OS and Mac OS X as an intuitive, user-friendly alternative to researchers with minimal prior coding experience to allow them to create publication-quality heatmaps using R graphics without sacrificing their desired level of customization. The simplicity of HeatmapGenerator is that it only requires the user to upload a preformatted input file and download the publicly available R software language, among a few other operating system-specific requirements.
Advanced features such as color, text labels, scaling, legend construction, and even database storage can be easily customized with no prior programming knowledge. Heatmaps are two-dimensional graphical false-color image representations of data using a user-defined color scheme to display values in a data matrix. In the medical research community, heatmaps are used to comparatively illustrate gene expression levels across a number of different samples (e.g., cells under different experimental conditions or samples from different patients) obtained from DNA microarray studies or high-throughput sequencing methods such as RNAseq. Genes that are upregulated (high relative expression value) are colored differently than genes that are downregulated (low relative expression value), thus providing a simultaneous visual representation of gene expression levels across multiple different samples. Heatmaps can be used to, for example, understand what groups of genes are turned on or turned off in various cancer samples as compared to control samples. For instance, heatmaps have been used for gene expression profiling of breast cancer samples to study the differential patterns of gene expression across multiple individual tumors.
There are a variety of publicly available and commercial graphical user interface (GUI) software for making heatmaps such as TM4 , GenePattern , Qlucore , and GENE-E. Although there is an abundance of such services, none of them boast comparable graphical capabilities such as those produced by the R programming language: popularly considered to be one of the most powerful and widely used statistical software among researchers in the medical community and the lingua franca for preparing heatmaps for publication. In contrast to previously released heatmap software, HeatmapGenerator is the first GUI heatmap software package capable of compiling heatmaps with just a few button clicks directly from the R language, without making users write a single line of code.
In other words, researchers can now use R to make heatmaps without needing any prior programming knowledge. Moreover, the GUI of HeatmapGenerator is the first software of its kind to employ a database storage system that stores a user’s previously generated heatmaps, in case the user wishes to go back to retrieve them from memory or compare them to other generated heatmaps, all within a central repository. Much of the publicly available heatmap software is not designed for handling gene expression data, instead focusing on an entirely unrelated niche such as geographical interactive online maps or even keyboard layout. Therefore, it is difficult for a medical researcher to find a suitable venue to produce biological heatmaps, instead having to rely on either outsourcing the data to a corporate venue or teaming up with a computational researcher well-versed in computer programming.
In this paper, we provide first-time publication of a user-friendly, R-centric GUI for the interactive real-time production of biological heatmaps in the R programming language via software development using the C programming language and the OpenGL application programming interface (API). A GUI has been developed to be the frontend to the HeatmapGenerator source code: an R program file designed to create a heatmap. The GUI is written in FLTK , a cross-platform object-oriented widget toolkit written in the C programming language with an interface to OpenGL. OpenGL is the industry standard for rendering cross-platform real-time 2D and 3D vector graphics with frequent interactive applications in flight simulation, scientific visualization, and computer-aided design. C is an efficient, compact, fast, portable programming language commonly used in safety-critical applications (e.g., aircraft control systems), operating system development (e.g., Apple Mac OS X, Microsoft Windows), and web search engines (e.g., Google) that links object-oriented programming, procedural programming, and generic programming in one holistic framework.
As such, HeatmapGenerator uses C to achieve maximal speed, efficiency, and stability when generating heatmap graphics. The source code that creates the heatmaps using the R programming language is executed by calling the Rscript batch interpreter that comes with a standard R download. HeatmapGenerator is able to execute the R heatmap source code within a GUI written in C/OpenGL using the FLTK framework so that the user can press the HeatmapGenerator user-interface buttons (e.g., to change the heatmap’s color scheme) while immediately visualizing these respective changes in the console window without needing to write a single line of code. R must be installed within the local user’s computer, but the GUI uses the R commands of the source code that plot the heatmap, all from a user-friendly clickable interface.
The source code utilizes the heatmap and heatmap.2 command within the gplots package of the R programming language to ultimately create the heatmap. For best use, we refer the reader to the operating system-specific “HeatmapGenerator manual.pdf” file that comes bundled with the HeatmapGenerator download. As stated in this file, to properly use the HeatmapGenerator software package, the user must first download R for (Mac)OSX/Windows at and preformat the input file to be a tab-delimited textfile (.txt) (an example is provided as EXAMPLE.txt in the HeatmapGenerator download).
Specifically, the input file should be a.txt file where each entry is separated by tabs, the first column should contain the item names (e.g., list of gene names), and the other columns should contain numbers corresponding to the respective items. An example of a.txt file table to be imported into HeatmapGenerator is shown in (refer to Figure ). Note that each column is labeled with its respective header (e.g., Gene Name, Control 1, Exp 1, etc.).
The HeatmapGenerator GUI design gives the user a full suite of options for creating, customizing, saving, storing, and choosing between simple and advanced heatmap construction. Running the HeatmapGenerator software package produces a variety of heatmaps (refer to Figure and Figure ) showing the relative expression levels of genes from either large or small datasets used as the input to the program. Simple input to the HeatmapGenerator can be as many as a few lines of a textfile up to thousands of lines of data in a textfile that are imported from a large array of analyses.
Some features of the GUI include a legend that is based on the data. The scaling parameters are developed based on the data that was supplied by the user. Also, the colors of the heatmap can be changed from simple buttons in order to specify a color scheme. After the user has run the HeatmapGenerator, the GUI is able to export the visualized heatmap in either.png or.jpg file formats that allow for publication-quality graphics. A.tiff image is always provided by the software by default. One additional feature is the ability to recall the previous heatmaps that were generated using a backend random access database. The user data is stored in a local database for easy recall and easy reusage to build the same heatmap in case the user wishes to reproduce the file.
A user can store an unlimited number of heatmaps in the database file system, provided that these heatmaps reside within the same folder as the HeatmapGenerator program itself (refer to Figure ). Figure 4 Database file system of HeatmapGenerator and software information. HeatmapGenerator can handle an unlimited number of heatmaps in its storage system. Software logo, licensure, copyright information, and user instructions are provided. Changing the color scheme through the simple click of a button changes the colors of the most downregulated and most upregulated genes portrayed in the heatmap. To change the font size of the labels (e.g., the gene names, experiment types) displayed on the heatmap produced, HeatmapGenerator automatically adjusts the size of text labels through its built-in scaling mechanisms. Hence, the user may easily specify modifications to the color selection, text labels, legends, scaling, as well as a local database for storage of previously used heatmaps.
Twomonusb mac twomonusb for mac. Windows 7 or later Mac OSX 10.9 or later. (Android Debug Bridge), You cannot use TwomonUSB for Android on Windows. If iPhone and iPad are in the environment that cannot synchronize with iTunes using USB, You cannot use TwomonUSB for iOS on Windows. Application that supports acceleration cannot be shown properly. Alternatives to TwomonUSB for iPhone, iPad, Mac, Windows, Android and more. Filter by license to discover only free or Open Source alternatives. This list contains a total of 7 apps similar to TwomonUSB.
Applications One use case for the HeatmapGenerator pertains to RNA next-generation sequencing data (RNAseq). The main goal of RNAseq is to quantify gene expression across multiple biological samples.
However, there is no gold standard analysis pipeline for RNAseq data. Rather, there are multiple forms of pipelines that look at the same data, but in two different ways. First, there is a very simplistic model of generating raw counts. These raw counts are generated after global alignments by a program such as HTseq. Second, there is an alternative mathematical approach to calculate a normalized value in order to scale the expression of each gene relative to its respective size (in base pairs).
Heat Map Generator Tutorial For Macbook Air
This method is performed by Cufflinks , which outputs the normalized value called fragments per kilobase per million (FPKM). The HeatmapGenerator is built to handle both types of data. The user can enter in either raw counts per gene or FPKM per gene, and the HeatmapGenerator will show the differential expression analyses in a visual manner. We provide an intuitive and user-friendly GUI software package, HeatmapGenerator, to create high-quality, customizable heatmaps generated using the popular data graphics capabilities of the R programming language. We achieve real-time visualization of these heatmaps using the industry standard high performance graphics of OpenGL. As such, instantaneous responsiveness in heatmap design is achieved through the graphical user interface interaction of a researcher’s input specifications. Furthermore, performance speed and software stability is achieved through the C programming language.
Moreover, a random access database structure is implemented to reliably store generated heatmaps. The R source code linked to C via an OpenGL API allows researchers the ability to create publication-quality biological heatmaps with minimal prior coding experience without sacrificing their desired level of customization: advanced features such as color, legends, text labels, scaling, and database storage can be easily customized with no prior programming knowledge.
Import Data Use the File menu to open a data file. See the file formats page for supported formats.
Example data files can be loaded by selecting FileOpen Example Data. Annotate Rows/Columns Annotate columns or rows based on entries provided in a tab delimited text file or an Excel.xls or.xlsx file. A colored bar visually identifies members of the same category. Annotatotions are primarily used for visualization.
In analyses such as marker selection, column annotations can also be used to identify phenotypes. To annotate columns/rows:. Create a tab delimited text file or an Excel.xls or.xlsx file. Select FileAnnotate Columns or FileAnnotate Rows and open the file created previously. GENE-E displays color bars below the column names or to the right of the row names to indicate the categories to which the columns/rows belong.
Select EditColumn Annotations or EditRow Annotations to edit the color for a category or to delete a category. New Heat Map To open a new heat map on a subset of your data:.
Select the desired columns and rows. Select ToolsNew Heat Map. Preferences Select GENE-EPreferences (Mac) or ViewOptions (other platforms) to modify the title, look and feel of the current visualization tool. GENE-E displays a window which provides options specific to the current visualization tool. Most options are self explanatory. The color tab controls the colors used in the heat map:.
Relative: GENE-E converts values to heat map colors using the mean and maximum values for each row or the standard deviations from the row mean for each row (as determined by the settings on this tab). GENE-E converts values to heat map colors using the minimum and maximum values in the entire data set (as determined by the settings on this tab). To change the color of the heat map click a colored square above the heat map legend and select a new color. Click and drag a colored square to move a control point. Click the add button to add a new control point. Click delete to delete an existing color. Sorting You can sort columns by column name, category, or annotation.
You can sort rows by row name, category, annotation, or the values in a particular column. To sort columns: Select ToolsSort Columns. Select the field(s) to sort.
Each drop-down list includes column (for column name) and all categories and annotations that you have loaded (in this example, the Phenotype category). To sort rows: Select ToolsSort Rows or click on a row header (shift-click to add a secondary sort). Select the field(s) to sort. Each drop-down list includes row (for row name), each column name, and all categories and annotations that you have loaded. Mask Data Masking rows or columns temporarily hides them from many GENE-E operations. For example, masked rows and columns can be omitted from new heat maps (ToolsNew Heat Map).
Highlight one or more rows or columns. Select ToolsMask Rows or ToolsMask Columns. Alternatively, right-click and select Mask Rows or Mask Columns from the context menu. You can clear masked columns/rows by highlighting one or more columns/rows and selecting ToolsUnmask Columns or ToolsUnmask Rows or to clear all masked columns/rows select ToolsClear Column Mask or ToolsClear Row Mask. Marker selection Marker selection identifies objects that are differentially expressed between two classes.
For each object, the analysis uses a test statistic to calculate the difference in expression between the classes and then estimates the significance (p-value) of the test score. It then corrects for multiple hypotheses testing (MHT) by computing both the false discovery rate (FDR) and the family-wise error rate (FWER). The output of marker selection consists of:. Score: The calculated value of the test statistic.
p-value: The estimated significance of the test statistic for this row (not yet corrected for MHT). p-value low: The estimated lower bound for the p-value. p-value high: The estimated upper bound for the p-value.
FDR(BH): The expected proportion of non-marker genes (false positives) within the set of genes declared to be differentially expressed. It is estimated using the Benjamini and Hochberg procedure. (Benjamini, Y. And Hochberg, Y.
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological). 1995.).
FWER: The probability of having any false positives. Hierarchical Clustering Hierarchical clustering recursively merges objects based on their pair-wise distance. Objects closest together are merged first, objects furthest apart are merged last.
The result is a tree structure, referred to as a dendogram, where the leaf nodes represent the original items and internal (higher) nodes represent the merges that occurred.